Datasets and Research Resources
Global biogeography of Ralstonia pathogens:
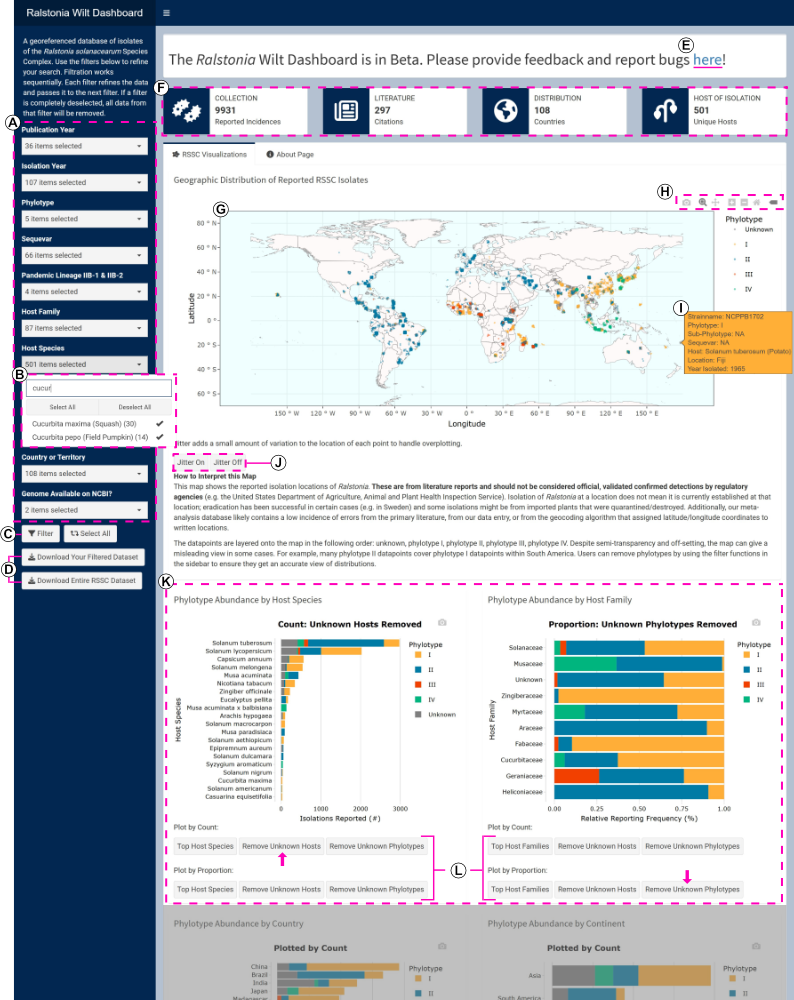
Seeking to unify the extensive literature on Ralstonia populations around the globe, we have an ongoing literature meta-analysis of Ralstonia solanacearum isolations (over 10,000 strains from over 300 papers). We publish updates to the Ralstonia isolation dataset as a “living” BioRxiv pre-print (Lowe-Power et al.).. In collaboration with Dr. Tessa Pierce-Ward, also adapted a web-based, interactive Dashboard that allows users to investigate patterns of Ralstonia biogeography, phylogeny, and host-of-isolation. Dashboard: https://ralstoniadashboard.shinyapps.io/RalstoniaWiltDashboard/ Our YouTube channel (linked below) has tutorials and case studies to demonstrate how to use the Dashboard to understand Ralstonia.
Lowe-Power Lab YouTube Channel
Seminar on Ralstonia biogeography and host range
Seminar on the physiological mechanisms that underly wilt symptoms during Ralstonia disease
FigShare
On FigShare, we release cartoons and raw data from papers. Most data is released with a CC-BY license that only requires attribution and does not restrict any reuse of the data.
KBase
We use KBase to generate organized datasets for comparative evolutionary genomics studies. We also have created public narratives that allow anyone to re-analyze Ralstonia RNAseq data from SRA.
We currate a list of useful KBase narratives on a page of our Protocols GitHub: https://github.com/lowepowerlab/protocols/blob/main/kbase.md
Interactive files to understand genetic organization of Ralstonia’s vgr-linked toxin/antitoxin clusters
These tools were built as part of our study:
-
A pangenomic atlas reveals that eco-evolutionary dynamics shape plant pathogen type VI secretion systems Nathalie Aoun, Stratton J. Georgoulis, Jason K. Avalos, Kimberly J. Grulla, Kasey Miqueo, Cloe Tom, Tiffany M. Lowe-Power bioRxiv 2023.09.05.556054; doi: https://doi.org/10.1101/2023.09.05.556054
-
There are not any recording of seminars on this study, but we have a slide deck available: https://figshare.com/articles/presentation/The_ecology_and_evolution_of_Ralstonia_s_toxin-secreting_molecular_weapon_T6SS_2023-08-18_ICPP_talk/23992947
Methods:
The interative synteny analysis files were created using Clinker. The documentation for Clinker can be found here
Reference: clinker & clustermap.js: Automatic generation of gene cluster comparison figures. Gilchrist, C.L.M., Chooi, Y.-H., 2020. Bioinformatics.
Synteny files organized by aux cluster
If you would like to re-analyze the files used in this dataset, the inputs are located in a repository on our FigShare (see link above)
- Interative HTML for aux1 (3 examples)
- Interative HTML for aux2 (4 examples)
- Interative HTML for aux3 (8 examples)
- Interative HTML for aux4 (7 examples)
- Interative HTML for aux5 (6 examples)
- Interative HTML for aux6 (9 examples)
- Interative HTML for aux7 (11 examples)
- Interative HTML for aux8 (6 examples)
- Interative HTML for aux9 (8 examples)
- Interative HTML for aux10 (5 examples)
- Interative HTML for aux11 (3 examples)
- Interative HTML for aux12 (4 examples)
- Interative HTML for aux13 (3 examples)
- Interative HTML for aux14-only (4 examples), aux20-only (3 examples), and combination aux14/aux20 (5 examples)
- Interative HTML for aux15 (4 examples) and aux18 (5 examples)
- Interative HTML for aux16 (5 examples)
- Interative HTML for aux17 (8 examples)
- Interative HTML for aux19 (2 examples)
- Interative HTML for aux21 (5 examples)
- Interative HTML for aux at tssK: aux10 (2 examples), aux17 (2 examples), aux22 (5 examples), aux23 (2 examples), aux24 (1 example) and a combination aux10/aux24 (1 example)
- Interative HTML for aux25 (3 examples)
Plasmid Maps
These plasmids are described in the linked publications. All plasmid maps are provided as read-only Benchling links. If you prefer a different cloning software, Benchlings allows exports as genbank files (.gb). If you find a broken link, or you’re looking for a vector sequence that I’ve overlooked, please email me
2018
Plasmids from: Metabolomics of tomato xylem sap during bacterial wilt reveals Ralstonia solanacearum produces abundant putrescine, a metabolite that accelerates wilt disease. Download PDF
Benchling: pKO-speC::Sm Benchling: pRCTspeC_com Benchling: pUFR80-hrcC
2017
Plasmids from: Plant-like bacterial expansins play contrasting roles in two tomato vascular pathogens. Download PDF
Benchling: pUFR80-RSc0818KO Benchling: pMiniTn7-RSc0818comp Benchling: pHN_RsExpA
2016
Plasmids from: Degradation of the plant defense signal salicylic acid protects *Ralstonia solanacearum from toxicity and enhances virulence on tobacco.* Download PDF
Benchling: pUFR80-KOnagGH Benchling: pST-KOnagAaGHAbIKL Benchling: pRCGnagGHcomp Benchling: pMiniTn7-nagAaGHAbIKL-comp Benchling: pMiniTn7-KRNP
Plasmids from: Functional identification of putrescine C- and N-hydroxylases. Download PDF
2015
Plasmids from: Hydroxycinnamic acid degradation, a broadly conserved trait, protects Ralstonia solanacearum from chemical plant defenses and contributes to root colonization and virulence. Download PDF
Benchling: pUFR80-fcsKO Benchling: pMiniTn7FcsComp
Parental Plasmids
Most of the plasmids I use frequently (e.g. pST-Blue, pUC18miniTn7t-Gm, & pRC* series for Ralstonia solanacearum GMI1000) are available online (PubMed or via manufacturers). However, I had to track down or reconstruct a few plasmid sequences. Those are listed below with original citations.
pUFR80 (Kan/sacB vector for markerless deletions)
Castañeda A, Reddy JD, El-Yacoubi B, Gabriel DW. 2005. Mutagenesis of all eight avr genes in Xanthomonas campestris pv. campestris had no detected effect on pathogenicity, but one avr gene affected race specificity. Mol Plant-Microbe Interact 18:1306-1317.
pAX1-Cm, pAX1-Em, & pAX1-Gm (Chromosomal complementation of Xylella Temecula1
Matsumoto, Young, and Igo. 2009. Chromosome-based genetic complementation system for Xylella fastidiosa. Appl Env Micro 75:1679-1687.