protocols
Lab protocols for the Lowe-Power lab (scroll down for table of contents Readme)
PCRs to detect & classify plant pathogenic Ralstonia
Writing/editing credit: Caitilyn Allen lab, Tiffany Lowe-Power
1. Universal plant pathogenic Ralstonia PCR (759/760)
Detects a region present in most R. solanacearum, R. pseudosolanacearum, R. syzygii strains (82/85 tested by Opina)
Citation: Opina et al. 1997. A Novel Method For Development Of Species And Strain-Specific DNA Probes And PCR Primers For Identifying Burkholderia solanacearum (Formerly Pseudomonas solanacearum). Asia Pacific Journal of Molecular Biology and Biotechnology Volume 5, No. I, April, 1997 pp. l9-30.)
| Primer Name | Sequence (5’ to 3’) |
|---|---|
| 759 | GTC GCC GTC AAC TCA CTT TCC |
| 760 | GTC GCC GTC AGC AAT GCG GAA TCG |
Product Size: 281 bp
PCR conditions
- Controls:
- No template control
- Positive control (e.g. gDNA from GMI1000)
- Calculate OneTaq or similar mastermix with pcr_workbook
- Thermal cycler conditions: Run OneTaq program with 58C anneal & 30 s extension time
- Resolve on 2% agarose gel
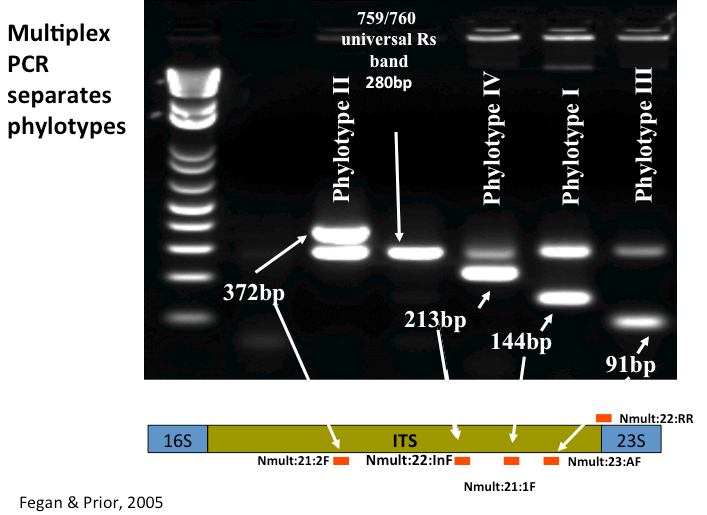
2. Phylotype PCR (Multiplex)
Citation: Fegan, M, and P. Prior 2005. How complex is the R. solanacearum species complex? In: Bacterial Wilt: The Disease and the Ralstonia solanacearum species complex (C. Allen, P. Prior, and A.C. Hayward, editors). APS Press, St Paul.
| Common Primers (CP) Name | Primer Name | Sequence (5’ to 3’) | Phylo specificity | Band size (bp)* |
|---|---|---|---|---|
| CP1 | Nmult21:1F | CGTTGATGAGGCGCGCAATTT | I | 144 |
| CP2 | Nmult21:2F | AAGTTATGGACGGTGGAAGTC | II | 372 |
| CP3 | ßNmult23:AF | ATTACSAGAGCAATCGAAAGATT | III | 91 |
| CP4 | Nmult22:InF | ATTGCCAAGACGAGAGAAGTA | IV | 213 |
| CP5 | Nmult22:RR | TCGCTTGACCCTATAACGAGTA | all | N/A |
PCR conditions
- Controls:
- No template control
- Positive control (ideally 1 strain from each phylotype, but at a minimum do a gDNA from a known isolate)
-
XXX Someone needs to add this to pcr_workbook Reactions were carried out in a total volume of 20 µl containing 1 x PCR buffer (supplied by the manufacturer of the polymerase) 1.5 mM MgCl2, 0.2 mM of each dNTP, 2U of Taq Polymerase 6 pmoles of the primers Nmult:21:1F, Nmult:21:2F, Nmult:22:InF, 18 pmoles of the primer Nmult:23:AF and 4 pmoles of the primers 759 and 760 (1). Reactions were heated to 96oC for 5 min and then cycled through 30 cycles of 94oC for 15s, 59oC for 30s and 72oC for 30s, followed by a final extension period of 10 min at 72oC.
- Thermal cycler conditions: Run OneTaq program with 59C anneal & 30 s extension time
- Resolve on 2% agarose gel
Expected results:
3. Detection of Race 3 Biovar 2 strains (U.S. Select Agent)
If reaction is positive, notify Tiffany Lowe-Power immediately. We are not permitted to receive, use, or store R3B2 organisms. If there is a positive reaction with the R3B2-specific primers, secure any materials with the R3B2 positive organism in a locked cabinet or freezer until they have been deactivated by autoclave. Tiffany will notify Aphis about the receipt of R3B2 organisms.
Citation: Opina et al. 1997. A Novel Method For Development Of Species And Strain-Specific DNA Probes And PCR Primers For Identifying Burkholderia solanacearum (Formerly Pseudomonas solanacearum). Asia Pacific Journal of Molecular Biology and Biotechnology Volume 5, No. I, April, 1997 pp. l9-30.)
| Primer Name | Sequence (5’ to 3’) |
|---|---|
| 630 | ATA CAG AAT TCG ACC GGC ACG |
| 631 | ATT CAC ATG CAA TTC GCC TAC |
Product Size: 307 bp
PCR conditions
- Controls:
- No template control
- Positive control (We do not have access to live Race 3 strains, but Caitilyn Allen’s lab generously purified DNA from UW551 for us)
- Calculate OneTaq or similar mastermix with pcr_workbook
- Thermal cycler conditions: Run OneTaq program with 60C anneal & 30 s extension time
- Resolve on 2% agarose gel
- If reaction is positive, notify Tiffany Lowe-Power immediately.
4. Multiplex PCR to Identify Ralstonia Species and Phylotype IIB1
Citation: “RSSC-Lineage Multiplex PCR” assay detects and differentiates Ralstonia solanacearum, R. pseudosolanacearum, R. syzygii and the R3bv2 subgroup, Sujan Paudel, 2022, https://doi.org/10.21203/rs.3.rs-1693987/v1
| Common Primers (CP) Name | Primer Name | Sequence (5’ to 3’) | Species Specificity | Band size (bp)* |
|---|---|---|---|---|
| CP6 | RssC-wF3 | TATATATCCTCGACTTTTCCATGAAGCTGTG | Ralstonia solanacearum species complex (F) | 162 |
| CP7 | RssC-wR3 | CTATATATATACCCCACTTGTTGAGGAACTG | Ralstonia solanacearum species complex (R) | 162 |
| CP8 | Rpseu-wF5 | TTTTATTTTTTTGGTGTCCGGGCCAAGATAG | R. pseudosolanacearum (Phylotype I and III) (F) | 251 |
| CP9 | Rpseu-wR5 | TTATATTACTCGAACGTGCTGCAAAACCACT | R. pseudosolanacearum (Phylotype I and III) (R) | 251 |
| CP10 | RsolP2-wF2 | ATTCTATTTATATCATGAGCGTTCCCCGACT | R. solanacearum (Phylotype II) (F) | 478 |
| CP11 | RsolP2-wR2 | TTTTTTTTTTTGAGGTAGCTGCTGGGGTTC | R. solanacearum (Phylotype II) (R) | 478 |
| CP12 | RsyzP4-wF4 | TTTCTTTTATTATAGTGTCGCGTCCGAACAG | R. syzygii (Phylotype IV) (F) | 664 |
| CP13 | RsyzP4-wR4 | TTTTTCTTTTTCGGTCTCTCCGTCTATCGTT | R. syzygii (Phylotype IV) (R) | 664 |
| CP14 | RsR3B2-wF | TTTCATTACTCATGACTGCAGAAACGCTTGA | Race 3 biovar 2 (II-R3bv2)(F) | 954 |
| CP15 | RsR3B2-wR | TATACATAACTATAGTTCGCCGTGCTCATCT | Race 3 biovar 2 (II-R3bv2) (R) | 954 |
5. Determine Sequevar of an Isolate
The sequevar system is a DNA based classification system of plant pathogenic Ralstonia. The system is based on DNA sequence alignments of a 750 bp region of the egl endoglucanase gene.
Step 1: Purify genomic DNA from the isolate
- Streak out the isolate on CPG+TZC and obtain single colonies.
- Inoculate a single colony into CPG broth and incubate overnight.
- Extract DNA from 1 ml of a dense culture using the DNeasy Blood & Tissue kit.
Step 2: Amplify a 750 bp region of the egl gene
| Primer Name | Sequence (5’ to 3’) |
|---|---|
| Endo-F | ATGCATGCCGCTGGTCGCCGC |
| Endo-R | GCGTTGCCCGGCACGAACACC |
Follow instructions for a High Fidelity PCR reaction (E.g. Kapa HiFi) to amplify the band. You will add 50 ng of purified genomic DNA as template. Run a program that is appropriate for a 750 bp target. Use an anneal temperature of 60 C (it looks like anything from 55-64C has been used in the literature).
Step 3: Visualize on an agarose gel to confirm a successful amplification
See the DNA electrophoresis protocol for more details.
- Make a 0.8% w/v agarose gel in 1x LAB buffer with ethidium bromide.
- Load 2 ul of your PCR reaction (mix with loading buffer). Don’t forget to run DNA ladder in a well.
- Run the gel
- Visualize the gel on a UV box. Save a photograph.
- Analyze the gel – annotate a few size markers on the ladder. You should have 1 strong, single band at 750 bp in your PCR wells.
Troubleshoot the PCR if necessary, otherwise proceed to step 4
Step 4: Purify the PCR reaction
Use the Zymo DNA clean & concentrator kit to purify your PCR products (1 prep per reaction).
Check DNA purity on the nanodrop. Record the estimated DNA concentration and 260/280 and 260/230 ratios. Also save a photograph of the full spectra of you DNA. Discuss with someone whether it looks “pure” or whether there are contaminant peaks.
Step 5: Submit the sample for Sanger Sequencing
Follow the Sanger Sequencing protocol. Sequence the reactions with both the Endo-F and Endo-R primers (separate reactions)
Step 6: Interpret the sequence.
Import the sequencing trace (.ab1 file) into Benchling in the “1-egl (partial) sequences”. Align the F and R and proofread the sequences.
After this point, follow the Cellier et al. reference protocol for infering sequevar. Interpret the results cautiously and know that some sequevars are polyphyletic on phylogenetic trees built with genomic data.